Introduction
AdaptiveFlow is a versatile, parallel workflow platform for carrying out virtual screening-related tasks on Linux-based computer clusters of any type and size which are managed by a batch system (such as SLURM).
Currently, there exist two versions of AdaptiveFlow, which are tailored to different types of tasks:
- AFLP: AdaptiveFlow for Ligand Preparation
- AFVS : AdaptiveFlow for Virtual Screenings
- AFU: AdaptiveFlow Unity (unifying ligand preparation and virtual screenings)
AFLP and AFVS use the same core technology regarding the workflow management and parallelization, and they can be used individually or in concert with each other. Additional versions are expected to arrive in the future.
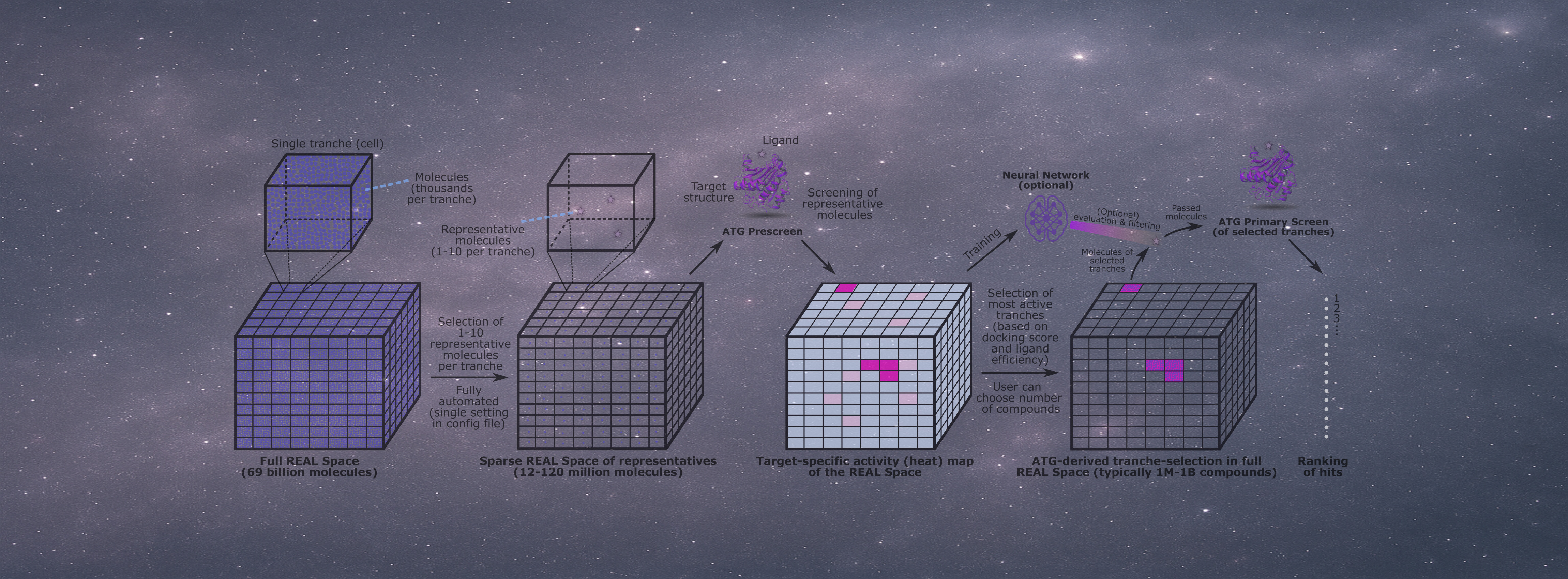
Pre-built, ready-to-dock ligand libraries for AFVS are available for free (in the download section).